You may switch between different representations of the microarray spot pseudoarray image. It may be viewed as several different types of pseudo images including an intensity gray value and a pseudo-color Red/Black/Green image for ratio (HP-X/HP-Y) and Zscore (HP-X - HP-Y) data. The p-Value results of comparing a HP-X 'set' with a HP-Y 'set' of samples, or the CV of the HP-EP 'list can be displayed as a color spectrum pseudoarray image.
Depending on the origin of the array data, it may have the same verisimilitude as the original arrays. Otherwise, it is displayed in a generic pseudoarray image containing grids that will fit the window - these are not the same as the original array image (see . However, the pseudoarrays are useful to getting a rough idea of the global changes in the data between arrays and how may genes pass the data filter.
When enabled using one of the commands in the Section 2.4.5 Clustering menu, cluster data appears as blue circles or squares drawn as overlays on the pseudoarray image. These options are discussed in the section on clustering. If you are doing clustering K-means clustering, the current cluster is displayed in the scatter plot if the latter is active.
Scatter plots, ratio and intensity histograms of the mean (HP-X/HP-Y) or (HP-X/HP-Y) 'set' data, or the F1/F2 or Cy3/Cy5 data. F1/F2 or Cy3/Cy5 plots are available if the data exists in your particular database. That might be the case with replicate spots or with Cy3/Cy5 data. If the normalization is set to a Zscore or log mean mode, it will compute Zscore scatter plots and histograms.
Clicking on spots in an array image or points in scatter plots sets
the current gene and will bring up data on the gene or (optionally)
access corresponding data from GenBank, UniGene, mAdb Clone, etc. databases in a
popup Web browser. Clicking on a bin in a ratio or intensity
histogram plot filters out all genes except for those in the range of
that bin.
Expression profiles plots of selected genes or subsets of genes for
all samples in the HP-E list. These are active plots with data reported
when the user clicks in the plot.
Clicking on a spot (i.e. gene) in the microarray pseudo image or on a
point (i.e. gene) in the scatter plot, it will define that gene as the
"current gene" that is used in other operations. The current gene is
indicated in both plots with a green circle
around it. Similarly, you may modify
the 'Edited Gene List' from either the pseudoarray image or the
scatter plot. When viewing is enabled, it overlays those genes with
magenta squares.
Figure 2.4.4 Plot menu - selecting Ratio Pseudoarray
image. This displays a pseudocolor show in the scale on the left that
indicates the ratio of the value of the HP-X sample / HP-Y sample (or
'sets' if the option to use HP-X and HP-Y 'sets' is enabled.) If The
data is Cy3/Cy5 data, then this displays the ratio of the ratios using
the current normalization. Various other pseudoarray image
representations could be used.
If the database that was loaded contains only one sample, the
pseudoarray image display defaults to the pseudograyscale spot
intensity mode. If there is at least one HP-X and one HP-Y sample,
then the Pseudocolor HP-X/Y ratio or Zdiff mode is the initial
default display. If there are duplicate spots for each gene, you may
generate a Pseudocolor F1F2 ratio or Zdiff mode image. If you
are using Cy3/Cy5 ratio data and the data is available as independent
channels for each HP, then you may plot Cy3 vs Cy5 for individual
samples.
When available on the database server, the original image may be
displayed in a separate popup Web browser.
Table 2.4.4.1. Pseudocolors assigned to spots to represent data in
the X/Y ratios or X-Y Zdiffs pseudocolor array images. Each color
represents the normalized X/Y ratio or X-Y Zdiff depending on
Normalization mode. The 9 colors of the boxes represent the normalized
expression ranges.
The same data is shown in a variety of normalization and display formats.
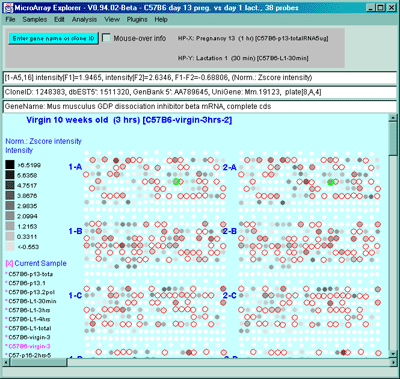
Figure 2.4.4.1.1.1 Pseudoarray intensity image of median normalized
intensities of the current HP sample (C57B6 virgin 10 weeks from MGAP
database). The graylevel scale on the left edge of the pseudoarray
image indicates the spot intensity. All pseudoarray images have scales
that vary depending on the type of pseudoarray being displayed.
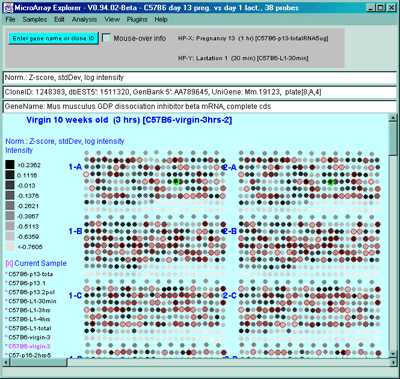
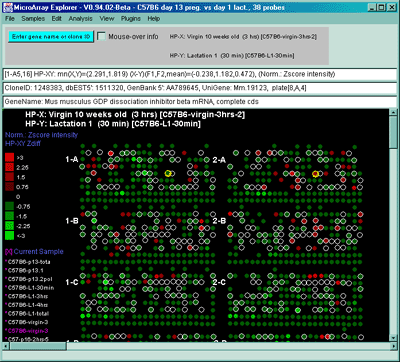
Figure 2.4.4.1.1.2 Pseudoarray intensity image of Zscore normalized
intensities of the current HP (C57B6 virgin 10 weeks from MGAP
database).
Figure 2.4.4.1.1.3 Pseudoarray intensity image of ZscoreLog normalized
intensities of the current HP (C57B6 virgin 10 weeks from MGAP database).
Figure 2.4.4.1.1.4 Pseudoarray intensity image of ZscoreLog
normalized intensities of the dual HP-X and HY-Y individual
samples. The Plot menu Show Microarray submenu toggle "Use dual
HP-X & HP-Y samples" option is set. HP-X is a C57B6 pregnancy day
13 and HP-Y is a Stat5a (-,-) pregnancy day 13.
Figure 2.4.4.1.1.5 Pseudoarray intensity image of ZscoreLog
normalized intensities of the dual HP-X and HY-Y sample 'sets'. The
Plot menu Show Microarray submenu toggle "Use dual HP-X & HP-Y
samples" option is set. The "Use HP-X & HP-Y 'sets' option in the
Samples menu. HP-X is the mean of three 'C57B6 pregnancy day 13' and
HP-Y is the mean of three 'Stat5a (-,-) pregnancy day 13'.
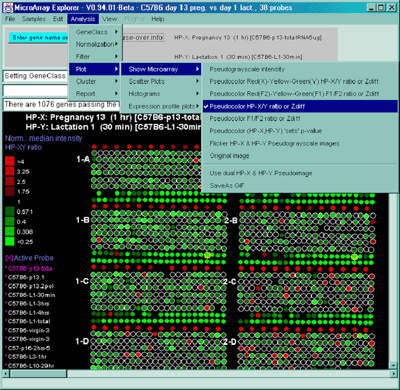
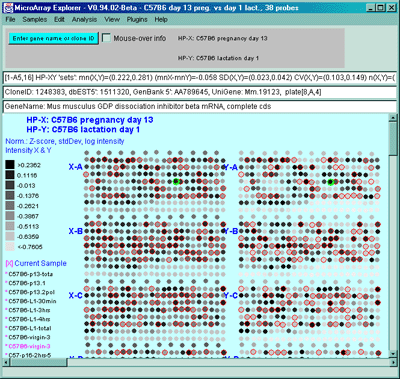
Figure 2.4.4.1.2.1 Pseudocolor array image of median normalized X/Y
ratios. HP-X is C57B6 pregnancy day 13 and HP-Y is Stat5a (-,-)
pregnancy day 13. Each spot's color represents the normalized X/Y
ratio depending on Normalization mode. The color of the box is one of
9 colors representing the normalized expression ranges and assigned
according to the table "Ratio
mode".
Figure 2.4.4.1.2.2 Pseudoarray color image of normalized X/Y 'set'
mean value ratios. Mean of three HP-X C57B5 pregnancy day 13
samples and mean of three HP-Y Stat5a (-,-) pregnancy day 13 samples.
Each spot's color represents the normalized X/Y 'set' ratios depending
on Normalization mode. The color of the box is one of 9 colors
representing the normalized expression ranges and assigned according
to the table "Ratio mode".
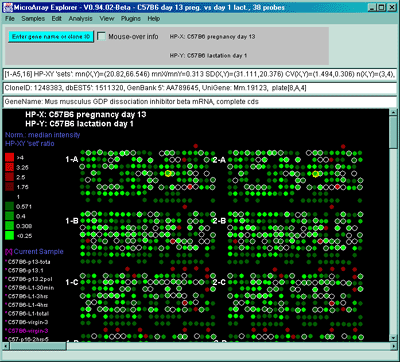
Figure 2.4.4.1.2.3 Pseudoarray color image of X-Y Zdiffs. HP-X
is C57B6 pregnancy day 13 and HP-Y is Stat5a (-,-) pregnancy day 13.
Each spot's color represents the normalized X-Y Zdiff depending using
the Zdiff normalization mode. The color of the box is one of 9 colors
representing the normalized expression ranges and assigned according
to the table "Zdiff mode".
Figure 2.4.4.1.2.4 Pseudoarray color image of X-Y Zdiff of log
data. HP-X C57B5 pregnancy day 13 sample and HP-Y Stat5a (-,-)
pregnancy day 13 sample. Each spot's color represents the normalized
X/Y ratio depending on ZdiffLog with StdDev normalization mode. The
color of the box is one of 9 colors representing the normalized
expression ranges and assigned according to the table "ZdiffLog mode".
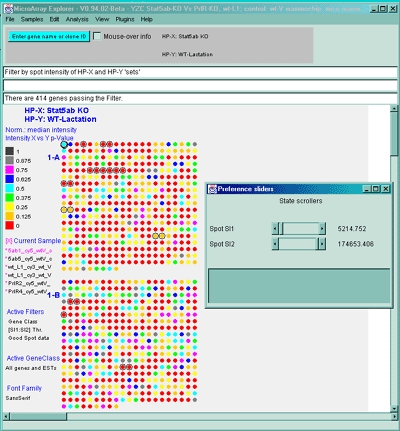
Figure 2.4.4.1.2.5 Pseudoarray image showing color-coded p-values
for t-test comparison of HP-X and HP-Y 'set' samples. The HP-X and
HP-Y sets both have 2 samples each (more is obviously much better).
The data was normalized using the Median and a spot intensity
[SI1:SI2] data filter was applied to eliminate some of the noisy data.
Each spot's color represents a p-value in the range indicated in the
scale in the left edge of the image. Note that although all spots are
assigned a p-Value, many may not be very significant because adequate
preprocessing of the data (such as normalization, and low intensity
spot removal, etc.). So use this display with care.
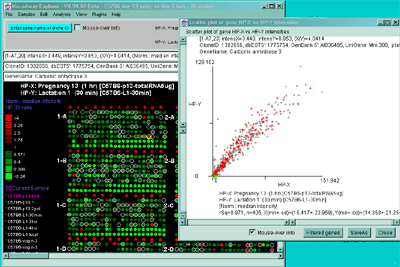
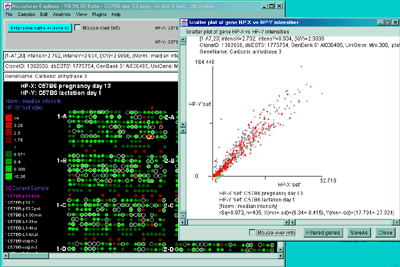
Figure 2.4.4.2 Scatter plot of HP-X and HP-Y single sample
data. HP-X is C57B6 pregnancy day 13 and HP-Y is pactation day 1.
A) An active scatter plot may be generated for the current HP-X
and HP-Y samples filtered by "All named genes". B) similar
plot for HP-X and HP-Y 'sets' of replicate samples (3 pregnancy and 4
lactation samples in the sets respectively). Clicking on a point in
the plot sets the current gene. C) Zoomed up region (of
B) at the bottom of the plot showing more detail and filtered
by just "All named genes". Zooming is performed by adjusting the X or
Y axes limits scroll bars. Note the points enclosed in magenta boxes
indicate genes in the E.G.L. gene list.
Figure 2.4.4.2.1 Scatter plot of multiple channel data from a
single sample. A) F1 Vs F2 data for a C57B6 pregnancy day 13
sample. B) Cy3 vs Cy5 data for a NCI mAdb mouse array sample.
C) Scatter plot of individual Cy3 channels from two different
ratio Cy3/Cy5 data hybridized samples. C) Scatter plot of
individual Cy3 channel of HP-X compared with Cy3 channel of HP-Y for
ratio Cy3/Cy5 data hybridized samples. D) Scatter plot of
individual Cy3 channel of HP-X compared with Cy5 channel of HP-Y for
ratio Cy3/Cy5 data hybridized samples.
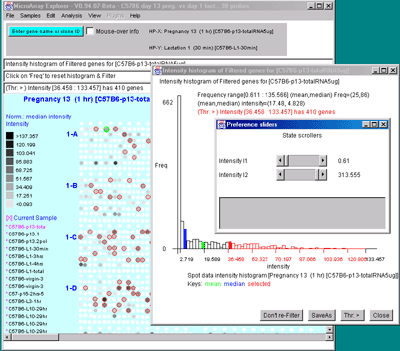
The Intensity selection plots a histogram of the gene intensity
data values for each Filtered spot (gene) in the current hybridized
sample.
The Histograms submenu includes:
If Cy3/Cy5 ratio data is being analyzed, then the F1F2 histogram menu
entry becomes
Figure 2.4.4.3 Histogram plots. A) Ratio histogram of
HP-X/HP-Y data with particular histogram bin selected with the
constraint set to filter all genes > that bin. HP-X is 13 day
pregnancy C57B6 and HP-Y is day 1 lactatation. The selected bin
thresholds are then used in the Filter with the resulting Filtered
genes shown in the array image. B) Zdiff histogram of HP-X -
HP-Y 'sets' for same data as (A) but with the ><
threshold constraint set to find genes outside of the symmetric
histogram range. C) Intensity histogram of HP-X data filtered
by [I1:I2] intensity range. As with ratio histograms, you can do
additional filtering by selecting a particular histogram bin that is
then used in the Filter. Filtering was disabled for the intensity
histogram. To apply the filter, the "Don't re-Filter" button would be
toggled to the "Re-Filter" state. The threshold constraints include:
=, >, <, >, <>, and ><. Note that each time
you click on the "Thr:" button, it cycles to the next option in the
threshold constraints list.
You many generate as many individual expression profile plots as you
want using the Display a gene's expr. profile for HP-E
command. However, only the last one will be active and will be updated
with different genes as you click on them in the microarray image
scatter plot. This could be used to compare the EP plots for several
different genes. First view the EP plot for one gene, then create a
new EP plot for the second gene, etc.
If you use the Display Filtered genes expr. profiles
for HP-E command, it will generate a scrollable list of
expression profile plots for all of the genes passing the Filter. If
the number of genes is very large, it may take a while.
You may interrogate a line corresponding to a particular HP sample in
a EP plot by moving the mouse over the line and then selecting the
line. This will cause the name of the HP, its intensity and CV to
appear in the plot. If the Err check box is set, then the mean
of the intensity is indicated by a short horizontal bar and the +- CV
by red vertical error bars above and below the mean. If the plot
style Line button is pressed, then the plot style is cycled
between Line (vertical lines for each point), Circle (small circles at
each point), and Curve (circles are connected). Pressing the button
repeatedly cycles through: Line (i.e. vertical vars),
Circle, or Curve (i.e. continuous curve of all
samples). In the case of mean expression profiles
used in K-means clustering, the standard deviation is used in
place of the CV value. The various clustering methods have EP
plots buttons. When they are invoked, the scrollable list of EP
plots is sorted by the clustering method ordered list of genes. This
enables you to view the data in the same order as that produced by the
cluster analysis. If the zoom nnX button is pressed,
then all of the plots are magnified by nn-fold to make low intensity
plots more visible. Pressing the button repeatedly cycles through:
1X, 2X, 5X, 10X and 20X. It does not change the data itself. The
Show HP names button pops up a numbered list of all HP entries
used in the expression profile. If you are in stand-alone mode, a
SaveAs GIF button will also be available for the EP overlay
mode (Figure 2.4.4.4.1) or individual EP plot. This saves the current
plot as a full resolution GIF file specified by the user in a popup
file browser window.
The Expression profile plots submenu contains:
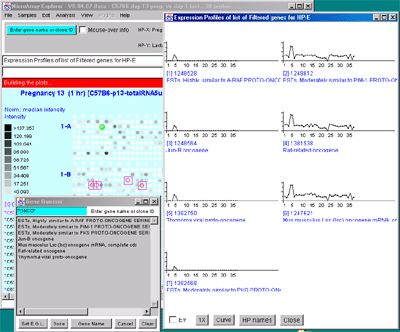
Figure 2.4.4.4 Expression profile plots. A) Individual
expression profile plots may be created by clicking on any
gene. Multiple instances may also be created. Here we show some of the
presentation options for the 38 sample MGAP database. Error bars are
computed for the standard error for that sample. There are three
different plotting options: line, circle and curve. #1 is the default
line plot with error bars. #2 is the line plot without the error bars
but clicking on line 7 to find out which sample it is and what the
intensity value is. #3 is the circle plot with error bars, and #4 is
the curve plot without error bars. Window #5 shows the list of samples
corresponding to the 38 points in the EP plots. B) List of
EPplots of the oncogenes and proto-oncogenes in the database (set by
the guesser with "onco" and "Set E.G.L." and the Edited Gene List
Filter). The list would become scrollable if there were more than 10
profiles. Setting the current gene would scroll the list to the EPplot
for the current gene.
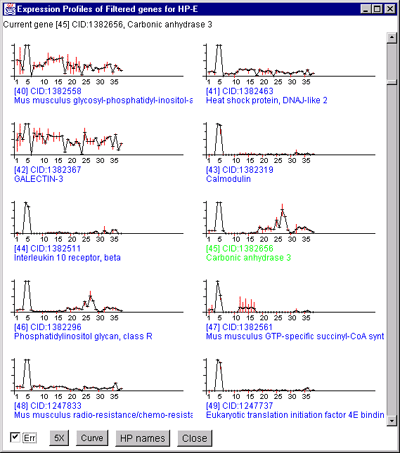
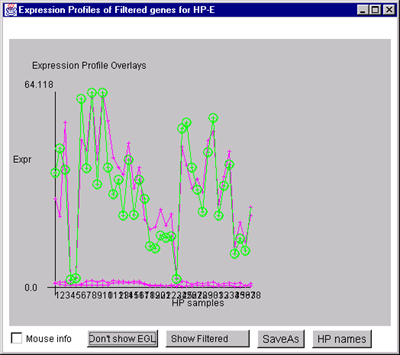
Figure 2.4.4.4.1 Expression profile plots.
A) Scrollable list of EP plots of Filtered named genes centered at
Carbonic anhydrase III.
B) Overlay plot of all named Filtered genes.
C) Overlay plot of all ONCO or PROTO-ONCO genes with the
draw EGL option active so the graphs are drawn for these genes.
2.4.4.1 Show microarray pseudoarray images menu
You may show the pseudoarray
image of the current hybridized samples using several
modalities. The grayscale pseudoarray image is generated from the
quantified spot data. If the data contains the actual spot positions
of the genes (as generated by the various array image quantification
program), the spots may be drawn using a scaled version of those
coordinates. Otherwise, a generic set of grids (and fields in there
are multiple fields) is synthesized to represent the spot
positions. Pseudoarray images may also be useful as an alternative
modality for displaying X/Y ratio or X-Y Zdiff data. If the
normalized intensities are the same, then the spot will appear as
black with the overall spot intensity depending on the spot
concentrations. High ratios and Zdiffs will be red and low values
green as shown in Table
2.4.4.1. The p-Value results of comparing a HP-X 'set' with a
HP-Y 'set' of samples can be displayed as a color spectrum pseudoarray
image.
![]() Pseudograyscale intensity [RB] - display a pseudograyscale
microarray pseudoarray image for the current H.P. where higher
intensity spots are mapped to black.
Pseudograyscale intensity [RB] - display a pseudograyscale
microarray pseudoarray image for the current H.P. where higher
intensity spots are mapped to black.
![]() Pseudocolor Red(X)-Yellow-Green(Y) HP-XY ratio or Zdiff [RB] -
display the HP-X and HP-Y microarrays as a pseudocolor image as
either a ratio (HP-X/HP-Y) or Zdiff (HP-X - HP-Y) if Zscore
normalization is in effect. The HP-X (red) and HP-Y (green)
components are added together so that high total
intensity values are yellow and low values are black. High
HP-X/HP-Y ratio values are more red and low values are more
green.
Pseudocolor Red(X)-Yellow-Green(Y) HP-XY ratio or Zdiff [RB] -
display the HP-X and HP-Y microarrays as a pseudocolor image as
either a ratio (HP-X/HP-Y) or Zdiff (HP-X - HP-Y) if Zscore
normalization is in effect. The HP-X (red) and HP-Y (green)
components are added together so that high total
intensity values are yellow and low values are black. High
HP-X/HP-Y ratio values are more red and low values are more
green.
![]() Pseudocolor Red(Cy5)-Yellow-Green(Cy3) Cy3/Cy5 data [RB] -
display the Cy3 and Cy5 microarray channels as a pseudocolor
array image. The the Cy5 (red) and Cy3 (green) components are
added together so that high total intensity values are
yellow and low values are black. High Cy3/Cy5 ratio values are
more green and low values are more red. Note: this is only
available with Cy3, Cy5 ratio data. Note: if the "Use ratio
median corrections" is enabled, the Cy3 channel is scaled using
the to the domain of the Cy5 channel by (median Cy5/median Cy3).
Pseudocolor Red(Cy5)-Yellow-Green(Cy3) Cy3/Cy5 data [RB] -
display the Cy3 and Cy5 microarray channels as a pseudocolor
array image. The the Cy5 (red) and Cy3 (green) components are
added together so that high total intensity values are
yellow and low values are black. High Cy3/Cy5 ratio values are
more green and low values are more red. Note: this is only
available with Cy3, Cy5 ratio data. Note: if the "Use ratio
median corrections" is enabled, the Cy3 channel is scaled using
the to the domain of the Cy5 channel by (median Cy5/median Cy3).
![]() Pseudocolor HP-X/Y ratio or Zdiff [RB] - display the HP-X and
HP-Y data as a pseudocolor array image as either a ratio
(HP-X/HP-Y) or Zdiff (HP-X - HP-Y) if Zscore normalization is in
effect. The high ratios or Zscores are red and low values are
green. Values in the middle are black.
Pseudocolor HP-X/Y ratio or Zdiff [RB] - display the HP-X and
HP-Y data as a pseudocolor array image as either a ratio
(HP-X/HP-Y) or Zdiff (HP-X - HP-Y) if Zscore normalization is in
effect. The high ratios or Zscores are red and low values are
green. Values in the middle are black.
![]() Pseudocolor (HP-X,HP-Y) 'sets' p-value [RB] - displays each
spot as a pseudocolor proportional to the p-Value in a t-Test of
the HP-X 'set' vs HP-Y 'set'. This can only be used when the
"Use HP 'sets' ..." option is enabled and there are at least 2
samples in both the HP-X 'set' and the HP-Y 'set'. Note that
unless proper normalization and filtering is used to remove poor
quality data, some of the p-Values will not be significant and
there will be a high false-positive rate. Use this display with
that in mind.
Pseudocolor (HP-X,HP-Y) 'sets' p-value [RB] - displays each
spot as a pseudocolor proportional to the p-Value in a t-Test of
the HP-X 'set' vs HP-Y 'set'. This can only be used when the
"Use HP 'sets' ..." option is enabled and there are at least 2
samples in both the HP-X 'set' and the HP-Y 'set'. Note that
unless proper normalization and filtering is used to remove poor
quality data, some of the p-Values will not be significant and
there will be a high false-positive rate. Use this display with
that in mind.
![]() Pseudocolor HP-EP 'list' CV (Coefficient of Variation)
[RB] - displays each spot as a pseudocolor proportional to
the CV of the spots within the subset of samples in the current
HP-EP 'list'. It is useful to look at the variation in a set of
replicate samples. Use the (Samples | Choose HP-X, HP-Y and HP-E
samples) command to define the HP-EP list of samples you want to
investigate.
Pseudocolor HP-EP 'list' CV (Coefficient of Variation)
[RB] - displays each spot as a pseudocolor proportional to
the CV of the spots within the subset of samples in the current
HP-EP 'list'. It is useful to look at the variation in a set of
replicate samples. Use the (Samples | Choose HP-X, HP-Y and HP-E
samples) command to define the HP-EP list of samples you want to
investigate.
![]() Pseudocolor Cy3/Cy5 (or F1/F2) ratio or Zdiff [RB] - display
the Cy3 and Cy5 (or F1 and F2 for duplicate spotted grids)
corresponding spots for the same genes as a pseudocolor array
image as either a ratio (Cy3/Cy5) or Zdiff (Cy3-Cy5) if Zscore
normalization is in effect. The high ratios or Zscores are red
and low values are green. Values in the middle are black.
Pseudocolor Cy3/Cy5 (or F1/F2) ratio or Zdiff [RB] - display
the Cy3 and Cy5 (or F1 and F2 for duplicate spotted grids)
corresponding spots for the same genes as a pseudocolor array
image as either a ratio (Cy3/Cy5) or Zdiff (Cy3-Cy5) if Zscore
normalization is in effect. The high ratios or Zscores are red
and low values are green. Values in the middle are black.
![]() Flicker HP-X & HP-Y Pseudoimages [RB] - toggle flickering the
HP-X and HP-Y pseudograyscale intensity array images.
Flicker HP-X & HP-Y Pseudoimages [RB] - toggle flickering the
HP-X and HP-Y pseudograyscale intensity array images.
![]() Use dual HP-X & HP-Y Pseudoimage [CB] - If have replicate
spots (F1,F2), toggle the display between F1 data in left grids
and F2 data in the right grids to mean HP-X data in the left
grids and HP-Y in the right grids for side by side comparisons.
Use dual HP-X & HP-Y Pseudoimage [CB] - If have replicate
spots (F1,F2), toggle the display between F1 data in left grids
and F2 data in the right grids to mean HP-X data in the left
grids and HP-Y in the right grids for side by side comparisons.
![]() "Scale pseudoarray image by
1/100 to zoom low-range values [CB] - rescales intensity and
(Cy3+Cy5) and (HP-X + HY-Y) (Red-Yellow-Green) plots to so
mid values are easier to visualize.
"Scale pseudoarray image by
1/100 to zoom low-range values [CB] - rescales intensity and
(Cy3+Cy5) and (HP-X + HY-Y) (Red-Yellow-Green) plots to so
mid values are easier to visualize.
.
.
.
.
.
.
.
.
.
Normalization
mode - RBG
bright green
.
.
dark green
Black
dark red
.
.
bright red
.
.
.
.
.
.
.
.
.
Normalization
mode - dichromasy
bright blue
.
.
dark blue
Black
dark orange
.
.
bright orange
<0.250X
0.307X
0.400X
0.571X
1.000X
1.75X
2.50X
3.25X
>4.00X
Ratio data
<-3.0
-2.25
-1.50
-0.75
0.00
0.75
1.50
2.75
>3.0
Zscore data
<-0.99
-0.742
-0.495
-0.247
0.000
0.247
0.495
0.742
>0.99
Zscore Log data
Clicking on a particular gene will report its specific quantification
and identification values (See
Section 3.3 on gene quantification). If the Enable display current
gene in popup genomic DB Web Browser option is set in the
View menu, then it will also pop up a Web browser with the
corresponding to the particular genomic DB data for that
database if it exists.2.4.4.1.1 Examples of microarray intensity data pseudoarray image
The relative intensity may be displayed for the current sample (last
HP-X or HP-Y selected) or two samples (HP-X or HP-Y samples or HP-X or
HP-Y 'sets' of samples). To show two samples side by side, enable the
Use dual HP-X & HP-Y Pseudoimage in the Show Microarray
submenu. To show averaged set data in the dual mode, enable the "Use
HP-X & HP-Y 'sets' option in the Samples menu. The grayscale
value reflects the current normalization mode.


2.4.4.1.2 Example of microarray ratio or Zdiff data pseudocolor image
The ratio (HP-X/HP-Y) or Zdiff HP-X - HP-Y) normalized intensity data
may be displayed as a pseudoarray image for HP-X and HP-Y, or HP-X and
HP-Y 'sets' of samples. To show averaged set data in the dual mode,
enable the "Use HP-X & HP-Y 'sets' option in the Samples menu.
The colors of the 9 scale boxes represent the normalized expression
ranges and is assigned according to the current normalization mode
listed in the table.


2.4.4.2 Scatter plots menu
Scatter plots include HP-X vs HP-Y intensity for comparing data
between HP-X and HP-Y samples (or sets if the HP-X and -Y sample
'sets' mode is enabled in the Samples menu - as is shown in Figure 2.2.1). You may
zoom into any area of the scatter plot as is shown in Figure
2.4.4.4.2(C). If there are duplicate spots for each gene, you may
plot F1 vs F2 intensity (or Cy3 vs Cy5 if using ratio data) for
comparing replicate data (or Cy3 and Cy5 ratio data channels) within
the same sample It will also compute the correlation coefficient for
the data and display it in the plot and in the message panel. The data
is the intensity values using the current normalization method. If you
are analyzing ratio Cy3/Cy5 data, you may compare Cy3 or Cy5 of the
HP-X sample against Cy3 or Cy5 of the HP-Y sample. If you are in
stand-alone mode, a SaveAs GIF button will also be
available. This saves the current plot as a full resolution GIF file
specified by the user in a popup file browser window.
rSq=0.974, n=1728, X(mn+-sd)=(4.477+-7.845), Y(mn+-sd)=(12.379+-24.810)
The Scatter plots submenu includes:

Scatter plots of data from multiple channels on the same sample
It is also possible to plot the separate channels within a single sample
against each other. For example F1 vs F2 in samples with replicate
data and Cy3 vs Cy5 in samples with separate ratio data channels.
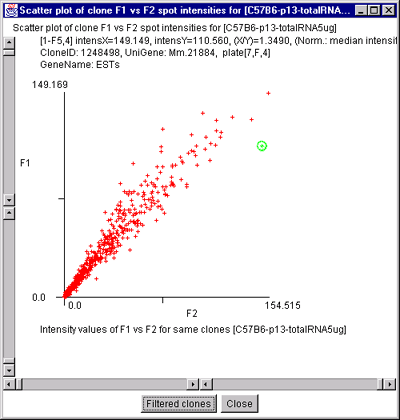

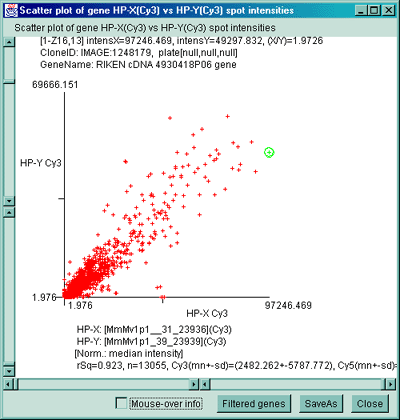

2.4.4.3 Histogram plots menu
You may compare ratios or Zdiffs of data using the HP-XY ratios or
Zdiff command to display a ratio histogram of Filtered intensity
data from two samples selected from the Samples menu. The HP-XY
'set' ratio or Zdiff is used if there are multiple samples in the
HP-X or HP-Y sets, then the mean values in each of the sets is used in
the calculations. If there are duplicate spots for each gene, you may
plot the F1F2 ratio or Zdiff histogram of the F1/F2 ratios or
F1-F2 Zdiff values for normalized data for each spot in the currently
displayed sample. If you are in stand-alone mode, a SaveAs GIF
button will also be available to save the current plot as a full
resolution GIF file specified by the user in a popup file browser
window.
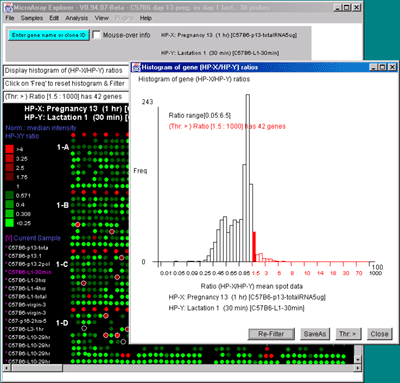
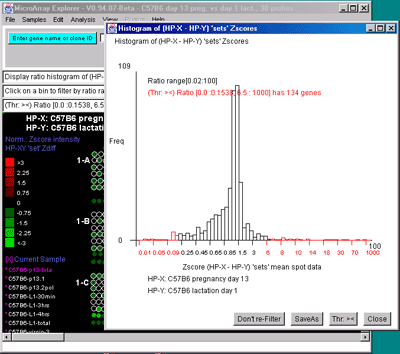
2.4.4.4 Expression profile plots menu
You may generate an individual expression profile plot (EP plot) or a
scrollable list of EP plots. The order list of hybridized samples to
plot are specified by the HP-E set. In the latter case, the genes are
specified by the data Filter.
![]() Use EP overlay else EP list [CB] - display Filtered genes
as an overlay plot of expression profiles, else as a scrollable
list of EP plots.
Use EP overlay else EP list [CB] - display Filtered genes
as an overlay plot of expression profiles, else as a scrollable
list of EP plots.