E.1 Internal data structures design to facilitate direct manipulation
MAExplorer was constructed using a number of fundamental data objects including clones (genes), hybridized samples (membranes or glass arrays), tables, etc. organized using an object-oriented methodology enforced by Java. Sets of genes are implemented as bit sets for efficiency in both storage and set-theoretic operations. With a set being implemented as 64-bits/word, a set intersection, union or difference can be performed on 64 genes in parallel in one logical (i.e. AND, OR, XOR) computer instruction. This makes the data filter quite efficient when computing the intersection of many gene sets. When ordered gene lists are required, memory and compute intensive lists are used - but only when needed. Tab-delimited ASCII is used as the basic I/O file type for all types of data. This simplifies I/O and allows data to be prepared with a variety of systems including Excel, array quantification programs, relational database systems, etc.
Another major decision was to use multiple pop-up windows for 2D
plots, histograms, expression profiles, clustergrams, reports, dialog
boxes, etc. rather than sharing a single window. These windows are
maintained by a special pop-up registry that handles many of the
bookkeeping chores involved with tracking and updating multiple
windows viewing the same underlying data. Whenever an event occurs
which may change the set of data filtered genes, the current gene or
the current cluster set of genes, the registry is notified. Some of
the events are the current clone changed, the Filter parameters
changed, the sample labels changed, the normalization method changed,
etc. It in turn notifies all relevant active plots, tables and
reports - requesting them to update themselves if necessary. This
object-oriented design greatly simplifies the process of synchronizing
the various data presentations with changes in the database.
A good intersection of the server-centric and client-centric methods
is to distribute the computation and data to the systems where they
can be handled most effectively. Because Java enables computation in
a Web browser, PCs currently available have enormous power and memory,
and high-speed Internet connections are readily available, it is now
possible to distribute some of the data and computations to the
desktop. If high-speed direct manipulation methodology is to be made
available on the Internet for microarray data mining, then it must be
brought to the user's desktop browser or local computer rather than
residing solely on the back-end server. This is the approach taken in
designing the MAExplorer.
Table E.2 Comparison of client-centric vs. server-centric data mining.
The table shows a comparison of some of the features of client-centric
and server-centric (using CGI and/or Applet) data mining analysis
methods. The client-centric approach presented here primarily uses
Java with data downloaded to the client's computer. A server-centric
approach might use a mix of HTML, CGI, servlet and Java. However, even
a client-centric approach may take advantage of server support for
additional functionality (e.g. accessing genomic servers to gain
additional information about specific genes or sets of genes).
E.2 Approaches to data mining: client-centric and server-centric models
There is a range of approaches for performing data mining of
microarray data over the Internet. However, all assume rapid access
to underlying databases and the ability to transform data from one
presentation mode to another where differences might be easily
observed. One extreme is the server-centric model using CGI or
Applets in Web browser. This assumes that all data search and analysis
is performed on a back-end server and graphic or tabular results from
the server are sent back to the researcher over the Internet. The
server-centric model has the advantage of keeping all user data
up-to-date, but the disadvantage of performing all computations and
graphics generation on the back-end server. Relying so much on the
server for major computations and graphics generation can result in
significant delays if the networks or servers are heavily loaded. The
other extreme is the client-centric model. Here all of the data being
analyzed is copied to a user's computer and computationally expensive
analyses are done there. This has the disadvantage for the user of
possibly not having the most up-to-date data to analyze as well as
setup time overhead. However, it does distribute the computational
load, allowing more effective data mining with many alternate views
and avoiding excessive delays during a data mining session. In both
the Web browser applet and the stand-alone application, data is
downloaded to MAExplorer. The difference being access to the local
file system with some additional capabilities in the case of the
latter.
Approach
Advantage (+)
disadvantage (-)Feature
Client-centric a)
+
Java programs run (pretty much) on all operating system
platforms as either stand-alone or applets (in browsers)
Client-centric b)
+
handles rapid response required for direct manipulation on
the new generation of very fast desktop computers
Client-centric c)
+
stand-alone version may be restarted quickly
from local data or data cached from the Web server
Client-centric d
+
size limitations are not a problem with
stand-alone Java applications
Client-centric e)
+
Java plug-ins allows prototyping new local and Web DB
analysis method functionality by any group of users
Client-centric f)
-
for the applet version, there is slow startup
because the program and all data has to be downloaded each time it is run
Client-centric g)
-
difficult to build large stable Web-applets handling very
large data sets. However, stand-alone applications don't have this problem
Client-centric h)
-
for the stand-alone application version,
it must be installed on client's computer where there nmight be some
level of incompatibility
Approach
Advantage (+)
disadvantage (-)Feature
Server-centric a)
+
may have better resources for very large data sets but with dependence on server
Server-centric b)
+
faster startup than downloaded applet since minimal GUI is required
and data does not have to be loaded before computation requests may be
made to the server
Server-centric c)
+
may be easier to prototype and distribute new functionality using third
party software such as RDBMS, S-plus, etc. using centralized CGI or
servlets where only one copy is required on the server
Server-centric d)
-
susceptible to Internet traffic bandwidth problems
for large numbers of users
Server-centric e)
-
susceptible to server-load dependencies for large numbers of users
Server-centric f)
-
difficult to get very rapid response for direct
manipulation for data mining
E.3 Conversion of microarray data files to MAExplorer format using
Cvt2Mae
A tool is being developed that converts microarray data files, both
commercial and one-of-a-kind research data to a complete MAExplorer
data format. Input data will be tab-delimited, although it may be possible
to use XML data at some point. When the tool becomes available, it will
be announced on the MAExplorer home page and in this manual.
Cvt2Mae data converter
Because it is difficult to manually edit user's microarray quantified
data files, we constructed the
Cvt2Mae data converter program (also see Appendix C.6). The idea
is to create array layouts for known array chips and to let the user
define their own for specialized arrays. These user-defined layouts
may then be saved and used in subsequent data conversions. The basic
problem of data conversion is that of "field picking" to map user data
fields to those required by MAExplorer, and of setting the appropriate
options in the MAExplorer configuration files. User-interactive
wizards query the user and then does this information to perform the
conversion generating the output data files that are ready to use with
MAExplorer. Cvt2Mae then generates the directory tree of required data
files described in Appendix C.
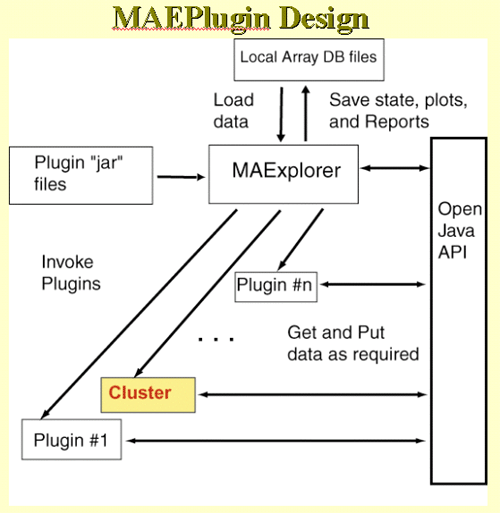
We are adding the ability for users to add their own Java Plug-in
Extensions to MAExplorer. These will extend capabilities of the core
MAExplorer program to other analysis methods by users. The MAEPlugins Web site will be an
Open Java API, open-source Java code examples, our plugins and donated
plugins, links to plugins at other Web sites. Typical plug-ins
include: normalization, Filters, PCA, clustering, client-server,
Web-server functional analysis of cluster results, etc. We group these
into three types of new analytic functionality:
![]() E.4 Extending MAExplorer functionality using Java Plugins
E.4 Extending MAExplorer functionality using Java Plugins
- Using Java code to implement the complete plugin. This means it will be portable across all systems.
- Accessing local programs written in any language ( eg. the R statistics package). This method may not be portable across all systems.
- Web-CGI or client-server to specialized genomic DBs Plug-ins. This method should be portable across all systems.
The following figures show the top level plugin design.
Figure E.4.1 Overall MAEPlugin design for MAExplorer. Plugins
are dynamically loaded into MAExplorer where they may be invoked from
a menu entry or by various other means such as startup, normalization,
etc.