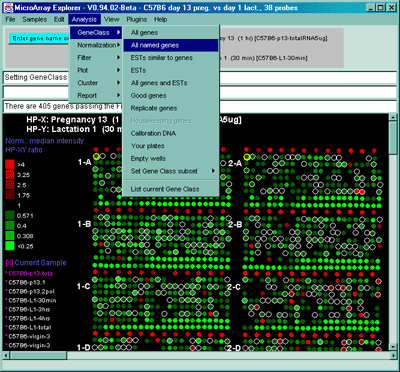
Figure 2.4.1.1 Gene Class menu. The user may select a subset of genes that belong to one of the classes of genes. This shows the user selecting the set of "All named genes" that are indicated with red (white) circle over the spots in the array intensity (ratio) pseudoarray image.
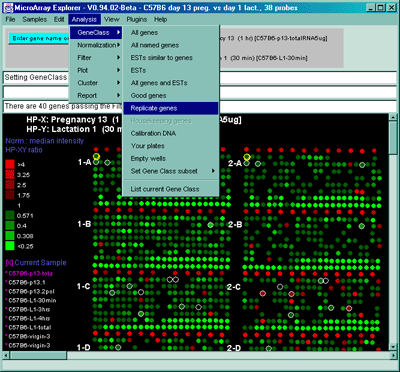
Figure 2.4.1.2 Example of all replicated genes occurring more than
once in the array. This was selected by using the GeneClass
'Replicate genes'. You may use the data Filter "Filter by genes with
replicates" instead of the GeneClass. This has the advantage that you
may use other GeneClasses (e.g. ESTs, or All named genes, etc.).
Alternatively, you can find all of the replicates for a particular
gene by 1) use the Gene Guesser to find the particular gene you want;
2) press "Set E.G.L." to save it as an Edited Gene List; 3) enable the
Filter "Filter by E.G.L." at the same. This will show all occurrences
of that gene.
The set of all genes constitutes a number of different gene
classes. It is possible to restrict the subsequent analysis to a
particular subset of these genes called a gene class. The
GeneClass menu operations include operations to select the
current set of genes to analyze from the set of all genes by their
membership in a gene class.
Some of the above gene classes are deduced from the gene name supplied
with the Gene In Plate Order (GIPO) file for the array. We use the
following automatic classification rules shown in Table 2.4.1.
Table 2.4.1 Rules for the automatic classification of gene names
into the default Gene Class sets. The gene name is analyzed
alphabetic-case independently.
![]() - select one of the gene class subsets [Future]
- select one of the gene class subsets [Future]
Gene class
Rule for class membership
All genes
all genes on the array
All named
genes not starting with "EST"
ESTs similar to genes
genes starting with "EST,"
ESTs
genes with the name "EST"
Replicate genes
genes with multiple copies
Calibration DNA
genes using the configuration file name "calibDNAname" (optional - see
Appendix Table C.4.1 )
Your plates
clones using the configuration file name "yourPlates" (optional - see
Appendix Table C.5.1-C))
Empty Wells empty wells where no spot exists
on the array indicated by keywords "empty", "empty well" or
"EmptyWell" (optional - see
Appendix Table C.5.1-C) )
Good Genes
spots on the array where the GIPO QualCheck data was used and was
valid. If it was not used, then it assumes all spots are good. (optional - see
Appendix Table C.4.1 )
2.4.1.1 GeneClass ontology subsets [Future]
If the Set Gene Class subset were activated, it might
include categories such as the following. If the categories exist and
the data is made available to MAExplorer, then it is possible to
specify gene subsets by Gene Class name. It is the responsibility of
the database creator to define a mapping table supporting these named
subsets of named genes.
2.4.1.2 Simulating Gene Class ontologies using Gene Set
operations
You can effectively implement finding ontology subsets for Gene Class
subsets using the following procedure. The trick is to repeatedly
define an E.G.L. gene subset using the gene name guesser to find the
genes of interest and save it as a named gene subset. Edit out genes
you don't want. Then you would repeatedly do the OR of gene sets of
interest, saving the result as a new named set. Then doing the OR of
another gene set with the set you just created, etc.
Procedure
