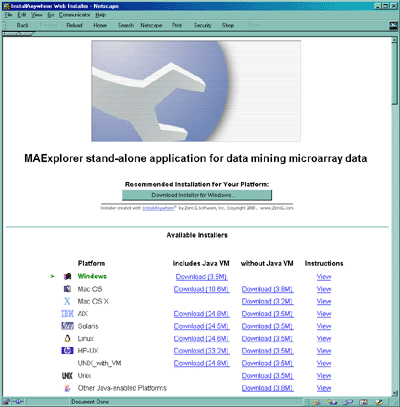
Figure D.1 Web page showing options for installing MAExplorer as a stand-alone application. Installers are available for Windows95/98/NT/2000/XP, Mac OS, Solaris, Linux, Unix, and other Java enabled platforms.
You first need to download MAExplorer for your particular type of
operating system. These include Windows 95/98/NT/2000/XP, MacOS for
Power PC, Sun Solaris, HP-UX, other Unix versions (e.g. Linux,
etc). The Windows, MacOS and Solaris versions include a Java run-time
(Java Virtual Machine) that works with MAExplorer. We recommend you
download the full distribution for your computer (which includes a
recent Java virtual machine (JVM) if it exists). This insures proper
operation of MAExplorer and does not interfere with other Java
applications you might have installed or will install.
This installation process uses a commercial "Java Installer" (InstallAnywhere(TM) by
ZeroG Inc.) that requests you "Grant" it permission to save the
installation on your computer. It will suggest where to install it or
you can install it wherever you want. For example, in Windows it may
suggest saving it in C:\Program Files\MAExplorer\ - you can
specify an alternative directory if the default disk does not have
much free space left.
D.2 Downloading MAExplorer for stand-alone use with other arrays
After MAExplorer is installed, you may start it by clicking on a
startup icon for one of the saved databases (Windows) or running the
program (Unix, Macintosh). In Windows, you may also start it from the
Window's "Start" menu. This stand-alone version enables you to save
data on your local machine and to run MAExplorer independent of an
Internet connection as well as other features that are discussed
throughout this manual.
D.2.2.1 Subsequent updating of only the MAExplorer JAR file from the
MAExplorer server
Generally, you can greatly shorten the subsequent download of new
versions of MAExplorer. This will work most of the time as most often
this is the only change in the distribution (not including the
documentation). If this causes problems, it may mean that the
other files have changed and you may have to reinstall the complete
distribution as in Appendix D.2 above.
Additional instructions for Installing MAExplorer on Sun Solaris
These instructions are valid for use with both TWM and CDE window
managers. If you have problems with the Sun installer, you may need to
update your Solaris OS system patches using a recent patch set.
It may involve more than a single patch. It is the latest Recommended
Patch Cluster from Sun. We STRONGLY recommend having
your System Admininistrator do this for you if you have not done this
before. Point your Web browser to:
http://sunsolve.Sun.COM/pub-cgi/show.pl?target=patches/patch-access
and choose the appropriate patch set for the version of Solaris (2.6,
7, or 8, etc.) that you are running. Do not choose any of the x86 versions
unless you are running Solaris x86. Click on either the Download HTTP
option or Download FTP option, and click the GO button to download the
patch set.
If you requested it, note that a Java virtual machine is included with
this download. It will run automatically when you run the shell
script.D.2.2 Enabling MAExplorer to fetch data from a microarray Web
server
When run as a stand-alone application, MAExplorer can be set up to
read data from a MAExplorer enabled Web database server and to cache
the data to your local computer. The following lists the three
variables that should be set in the .mae startup database
file (see full
table in discussion on setting up the configuration file). These
variables may be set from the Edit menu Preferences submenu Use Web
DB and Web DB data caching toggle switches, and the URL of
the database by the Set Web DB command.
