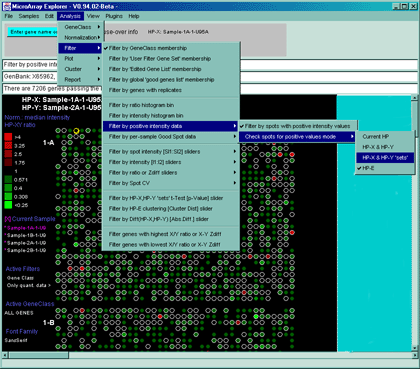
Figure 2.4.3 Filter menu. The Filter menu is a cascade of data filters that restrict the set of genes passing all filters that have been enabled and whatever the criteria was that was set for those filters. This figure shows the GeneClass filter set to "All genes and ESTs", the spot CV filter and Ratio (X/Y) range filters being set interactively by the scroll bars on the right. The genes that pass the filter are indicated with a red (white) circle in the array intensity (ratio) pseudoarray image.
The Filter menu options are used to restrict the set of genes by pre-filtering the data with a series of cascaded filter criteria and tests. The resulting subset of genes passing the filter are then used in the plots, reports and other data analysis methods. Some of the filters require additional parameters that are set by the State scrollers. The user will automatically be prompted for changes to these scollers (a threshold scrollers window will pop up) when the filter is activated or change. These values may also be set from the Adjust all Filter threshold scrollers entry in the Preferences submenu in the Edit menu. The filters are broken up into subgroups in the following menu with the grouping haveing more to do with the criteria (i.e. gene set membership, data range, or statistical tests).
-
 Filter by GeneClass membership [CB] - only include genes
that are members of the current GeneClass.
Filter by GeneClass membership [CB] - only include genes
that are members of the current GeneClass.
-
 Filter by 'User Filter Gene Set' membership [CB] - only
include genes that are members of the current 'User Filter Gene
Set'.
Filter by 'User Filter Gene Set' membership [CB] - only
include genes that are members of the current 'User Filter Gene
Set'.
-
 Filter by 'Edited Genes List' membership [CB] - only include
genes that are members of the 'Edited Gene List'.
Filter by 'Edited Genes List' membership [CB] - only include
genes that are members of the 'Edited Gene List'.
-
 Filter by global 'Good Genes List' membership [CB] - only
include genes that are members of the list of good genes. [These
genes are indentified by a QualCheck entry in the GIPO database
file.]
Filter by global 'Good Genes List' membership [CB] - only
include genes that are members of the list of good genes. [These
genes are indentified by a QualCheck entry in the GIPO database
file.]
-
 Filter by 'Genes with replicates' [CB] - only include
genes that genes that have at least 2 copies of the gene
replicated on the array. Note: duplicated
genes (i.e. F1, F2, etc) are not considered replicates
for this purpose.
Filter by 'Genes with replicates' [CB] - only include
genes that genes that have at least 2 copies of the gene
replicated on the array. Note: duplicated
genes (i.e. F1, F2, etc) are not considered replicates
for this purpose.
- -------------------
-
 Filter by ratio or Zdiff histogram bin [CB] - only include
genes that are in the range of the ratio or Zdiff histogram
bin you have clicked on (should be set from histogram plot, but may be
turned off here)
Filter by ratio or Zdiff histogram bin [CB] - only include
genes that are in the range of the ratio or Zdiff histogram
bin you have clicked on (should be set from histogram plot, but may be
turned off here)
 Filter by intensity or (Cy3/Cy5) histogram bin [CB] - only
include genes that are in the range of the intensity histogram
bin you have clicked on (should be set from histogram plot, but may be
turned off here)
Filter by intensity or (Cy3/Cy5) histogram bin [CB] - only
include genes that are in the range of the intensity histogram
bin you have clicked on (should be set from histogram plot, but may be
turned off here)
- Filter by positive intensity data
 - filter by positive intensity data
if the data may contain negative numbers. Otherwise it will use
both positive and negative data. If the database has 2 channels
(F1, F2) or (Cy3,Cy5) each channel is checked. If the background
correction is enabled, the background corrected values are
tested to see if any of them are negative.
- filter by positive intensity data
if the data may contain negative numbers. Otherwise it will use
both positive and negative data. If the database has 2 channels
(F1, F2) or (Cy3,Cy5) each channel is checked. If the background
correction is enabled, the background corrected values are
tested to see if any of them are negative.
 Filter by genes with non-zero intensity [CB] - only
include genes that have non-zero density. This protects against
zero data that may be present in the database when taking logs of
the data.
Filter by genes with non-zero intensity [CB] - only
include genes that have non-zero density. This protects against
zero data that may be present in the database when taking logs of
the data.
- Filter by per-sample Good Spot data
 - filter out genes that do
not have "Good Spot" values (defined by the optional QualCheck
spot data on a per-sample (i.e. HP) basis. See the list of
codes in Appendix
C.4). If there is no such spot quality data, then all spots
are considered "good".
- filter out genes that do
not have "Good Spot" values (defined by the optional QualCheck
spot data on a per-sample (i.e. HP) basis. See the list of
codes in Appendix
C.4). If there is no such spot quality data, then all spots
are considered "good".
- Filter by per-sample Spot Detection Value data
 - filter out genes that do not have
"Detection Value" values (defined by the optional DetValue or
CorrCoef spot data on a per-sample (i.e. HP) basis. Typical
Detection Values could be the Affymetrix MAS5.0 "Detection
p-value" or other continuous value of spot detection quality.
- filter out genes that do not have
"Detection Value" values (defined by the optional DetValue or
CorrCoef spot data on a per-sample (i.e. HP) basis. Typical
Detection Values could be the Affymetrix MAS5.0 "Detection
p-value" or other continuous value of spot detection quality.
- -------------------
- Filter by spot intensity [SI1:SI2] sliders
 - filter by individual spot intensity
(Cy3 and Cy5 channels if ratio data) within [SI1:SI2] threshold
ranges
- filter by individual spot intensity
(Cy3 and Cy5 channels if ratio data) within [SI1:SI2] threshold
ranges
- Filter by [I1:I2] sliders
 - filter by gene expression (or
Cy3/Cy5 if ratio data) within [I1:I2] threshold ranges
- filter by gene expression (or
Cy3/Cy5 if ratio data) within [I1:I2] threshold ranges
- Filter by ratio or Zdiff sliders
 - filter by gene ratios or Zdiff values
within [R1:R2] or [Z1:Z2] threshold ranges (depending on the
normalization method)
- filter by gene ratios or Zdiff values
within [R1:R2] or [Z1:Z2] threshold ranges (depending on the
normalization method)
- Filter by Cy3/Cy5 HP-X ratio or Zdiff sliders
 - filter by gene ratios or Zdiff values
within [CR1:CR2] or [CZ1:CZ2] threshold ranges (depending on the
normalization method). This is useful for filtering data from a
single sample.
- filter by gene ratios or Zdiff values
within [CR1:CR2] or [CZ1:CZ2] threshold ranges (depending on the
normalization method). This is useful for filtering data from a
single sample.
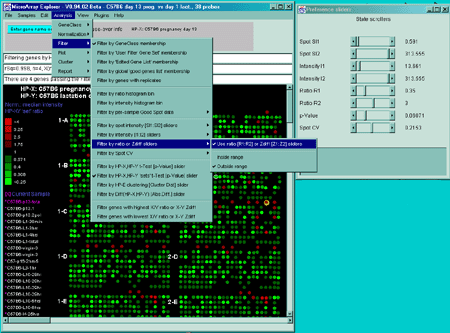
- Filter by spot CV
 - filter
out genes that do not meet minimum Coefficient of Variation
(CV) values of spot replicates (F1 and F2 for the same HP,
replicates in HP-X and HP-Y 'sets' of samples etc.).
- filter
out genes that do not meet minimum Coefficient of Variation
(CV) values of spot replicates (F1 and F2 for the same HP,
replicates in HP-X and HP-Y 'sets' of samples etc.).
- -------------------
 Filter by HP-X,HP-Y t-Test
[p-value] slider [RB] - only include genes that meet the
HP-X,HP-Y t-Test criteria if they have (F1,F2) duplicate spot
(this is a weak form of the t-Test).
Filter by HP-X,HP-Y t-Test
[p-value] slider [RB] - only include genes that meet the
HP-X,HP-Y t-Test criteria if they have (F1,F2) duplicate spot
(this is a weak form of the t-Test).
 Filter by HP-X,HP-Y 'sets'
t-Test [p-value] slider [RB] - only include genes that meet the
HP-X,HP-Y 'sets' t-Test criteria (only works if using HP-X and
HP-Y 'sets' mode where there are replicate samples).
Filter by HP-X,HP-Y 'sets'
t-Test [p-value] slider [RB] - only include genes that meet the
HP-X,HP-Y 'sets' t-Test criteria (only works if using HP-X and
HP-Y 'sets' mode where there are replicate samples).
 Filter by HP-X,HP-Y 'sets'
Kolmogorov-Smirnov test [p-value] slider [RB] - only include
genes that meet the HP-X,HP-Y 'sets' KS-Test criteria (only
works if using HP-X and HP-Y 'sets' mode where there are
replicate samples).
Filter by HP-X,HP-Y 'sets'
Kolmogorov-Smirnov test [p-value] slider [RB] - only include
genes that meet the HP-X,HP-Y 'sets' KS-Test criteria (only
works if using HP-X and HP-Y 'sets' mode where there are
replicate samples).
 Filter by current Ordered
Condition List (OCL) F-Test [p-Value] slider [RB] - only
include genes that meet the F-test criteria on the current
OCL. This only works if there are at least 2 (replicate)
samples/condition for each of the condition sets in the OCL.
See info on defining
the OCL and
using the OCL data.
Filter by current Ordered
Condition List (OCL) F-Test [p-Value] slider [RB] - only
include genes that meet the F-test criteria on the current
OCL. This only works if there are at least 2 (replicate)
samples/condition for each of the condition sets in the OCL.
See info on defining
the OCL and
using the OCL data.
- -------------------
 Filter by HP-E clustering
[Cluster dist] slider [CB] - only include genes that meet the
clustering criteria (alternatively, see the Cluster menu
commands).
Filter by HP-E clustering
[Cluster dist] slider [CB] - only include genes that meet the
clustering criteria (alternatively, see the Cluster menu
commands).
 Filter by Diff(HP-X,HP-Y) [Abs.Diff.] slider [CB] - only
include genes whose absolute difference between mean HP-X and
HP-Y (single or 'sets') is < threshold.
Filter by Diff(HP-X,HP-Y) [Abs.Diff.] slider [CB] - only
include genes whose absolute difference between mean HP-X and
HP-Y (single or 'sets') is < threshold.
- -------------------
 Filter N genes with highest
X/Y ratio or X-Y Zdiff [CB] - look at highest ratios or Zdiff
values. The value of N is set in the Edit menu preferences.
Filter N genes with highest
X/Y ratio or X-Y Zdiff [CB] - look at highest ratios or Zdiff
values. The value of N is set in the Edit menu preferences.
 Filter N genes with lowest
X/Y ratio or X-Y Zdiff [CB] - look at lowest ratios. The value of
N is set in preferences. N is set in the Edit menu preferences.
Filter N genes with lowest
X/Y ratio or X-Y Zdiff [CB] - look at lowest ratios. The value of
N is set in preferences. N is set in the Edit menu preferences.
The Filter by positive intensity
data submenu filter contains options that specify which spot
intensity values are to be considered when excluding negative
quantified spot data. Note: this filter only makes sense if your data
might have negative values (e.g. Affymetrix chip "Avg Diff" data) or a
background corrected value that is less than 0.0. The filter is
enabled by setting the "Filter by spots with positive intensity"
checkbox. Negative intensity values may occur with some types of
arrays quantification programs. In the "Check spots for positive
values mode" submenu, you may set the samples where the test may be
applied to spots from the current HP, the single (HP-X,HP-Y) samples,
(HP-X,HP-Y) 'sets' (replicated spots), or samples in the HP-E list
selected to be used in the filter. If there are (F1,F2) or (Cy3/Cy5)
data, then each spot must meet the threshold criteria.
The Filter by Good Spot data submenu filter contains options
that specify spots based on their quality. It filters out genes that
have that do not have "Good Spot" values defined by the optional
QualCheck spot data. (See the list of codes in Appendix C.4). If there is no
such spot quality data, then all spots are considered "good". The
filter is enabled by setting the "Filter by spots with Good Spot
values" checkbox. All spots for the specified samples must meet the
criteria. In the "Check spots for Good Spot mode" submenu, you may set
the samples where the test may be applied to spots from the current
HP, the single (HP-X,HP-Y) samples, (HP-X,HP-Y) 'sets' (replicated
spots), or samples in the HP-E list selected to be used in the filter.
The Filter by Spot Detection Value data submenu filter
contains options that specify spots based on their spot detection
value quality metric over the range of [0.0 : 1.0]. The filter is
available only if the data exists for your database and is ignored
otherwise. If active, it pops up a "Spot Detection Value" slider in
the range of [0.0 : 1.0]. Only spots greater than the slider value
pass the filter. This data could be the Affymetrix MAS5.0 "Detection
p-value" or some other metric correlated with spot detection quality.
The filter is enabled by setting the "Filter by per-sample Spot
Detection Value" checkbox. All spots for the specified samples must
meet the criteria. In the "Check spots for Spot Detection Value mode"
submenu, you may set the samples where the test may be applied to
spots from the current HP, the single (HP-X,HP-Y) samples, (HP-X,HP-Y)
'sets' (replicated spots), or samples in the HP-E list selected to be
used in the filter.
The Filter by spot intensity [SI1:SI2] sliders submenu contains
options that determines how individual spot intensity thresholding is
to be applied in the Filter.
The Filter by [I1:I2] sliders submenu contains options that
determines how spot expression (intensity or (Cy3/Cy5) ratio value)
thresholding is to be applied in the Filter:
The Filter by ratio or Zdiff sliders submenu contains options
that determines how spot-ratio thresholding is to be applied in the
Filter. The spot ratio is mean HP-X / mean HP-Y for sets of
samples. The spot Zdiff is used if one of the Zscore normalization
methods is active and is computed as (mean HP-X - mean HP-Y) for sets
of samples.
The Filter by Cy3/Cy5 HP-X ratio or Zdiff sliders submenu
contains options that determines how spot Cy3/Cy5 HP-X ratio
thresholding is to be applied in the Filter. The spot ratio is Cy3/Cy5
for normalized data unless one of the Zscore methods is used. In that
case, the Zdiff is used and is computed as (Cy3 - Cy5) for sets of
samples. If HP-X 'sets' is used, then it computes the mean Cy3 value
and the mean Cy5 value and uses those values in the above
computations.
The Filter by spot CV submenu filter contains options that
specify how the Coefficient Of Variation of the (F1,F2) or (HP-X,HP-Y)
'sets' (replicated spots) is to be used in the filter. The (F1,F2) CV
is available only if there are duplicate spots on the HPs.
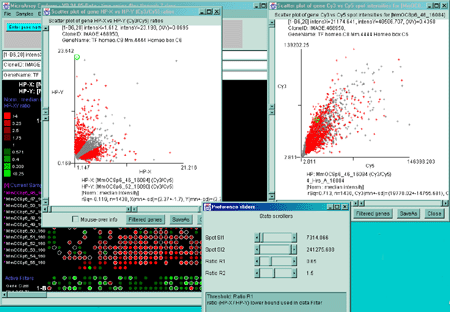
Figure 2.4.3.1 Filtering using multiple scrollers. This example
is of Cy3/Cy5 time series data. It filters normalized spot intensity
of the Cy3 and Cy5 channels independently ([SI1:SI2] inside range)
where low intensity spots are eliminated. It then filters out genes
outside of the [R1:R2] ratio range.
Figure 2.4.3.2 Using the Positive Intensity data Filter.
This allows removing negative data if the data contains negative
intensity values (e.g. Some Affymetrix data has negative Average Difference
values which could be read as Intensity for MAExplorer).
![]() Current HP [RB] - spots in current sample spots
Current HP [RB] - spots in current sample spots
![]() HP-X & HP-Y [RB] - spots in X and Y single samples
HP-X & HP-Y [RB] - spots in X and Y single samples
![]() HP-X or HP-Y 'sets' [RB] - spots in the HP-X set or HP-Y set
HP-X or HP-Y 'sets' [RB] - spots in the HP-X set or HP-Y set
![]() HP-X & HP-Y 'sets' [RB] - spots in both the HP-X set and
HP-Y set
HP-X & HP-Y 'sets' [RB] - spots in both the HP-X set and
HP-Y set
![]() HP-E [RB] - spots in HPs in expression profile list
HP-E [RB] - spots in HPs in expression profile list
![]() Current HP [RB] - spots in current sample spots
Current HP [RB] - spots in current sample spots
![]() HP-X and HP-Y [RB] - spots in X and Y single samples
HP-X and HP-Y [RB] - spots in X and Y single samples
![]() HP-X or HP-Y 'sets' [RB] - spots in HP-X set or HP-Y set
HP-X or HP-Y 'sets' [RB] - spots in HP-X set or HP-Y set
![]() HP-X and HP-Y 'sets' [RB] - spots in HP-X set and HP-Y set
HP-X and HP-Y 'sets' [RB] - spots in HP-X set and HP-Y set
![]() HP-E [RB] - spots in HPs in expression profile list
HP-E [RB] - spots in HPs in expression profile list
![]() Current HP [RB] - spots in current sample spots
Current HP [RB] - spots in current sample spots
![]() HP-X and HP-Y [RB] - spots in X and Y single samples
HP-X and HP-Y [RB] - spots in X and Y single samples
![]() HP-X or HP-Y 'sets' [RB] - spots in HP-X set or HP-Y set
HP-X or HP-Y 'sets' [RB] - spots in HP-X set or HP-Y set
![]() HP-X and HP-Y 'sets' [RB] - spots in HP-X set and HP-Y set
HP-X and HP-Y 'sets' [RB] - spots in HP-X set and HP-Y set
![]() HP-E [RB] - spots in HPs in expression profile list
HP-E [RB] - spots in HPs in expression profile list
![]() Use spot intensity [SI1:SI2] sliders [CB] - use spot
intensity thresholding
Use spot intensity [SI1:SI2] sliders [CB] - use spot
intensity thresholding
![]() Inside [RB] - test inside of [SI1:SI2] range
Inside [RB] - test inside of [SI1:SI2] range
![]() Outside [RB] - test outside of [SI1:SI2] range
Outside [RB] - test outside of [SI1:SI2] range
![]() -
specify which samples are tested
-
specify which samples are tested
![]() -
specify which additional constraints are used. This is useful
for finding genes with high or low expression but that has some
samples that have opposite expression.
-
specify which additional constraints are used. This is useful
for finding genes with high or low expression but that has some
samples that have opposite expression.
![]() Current HP [RB] - spots in current sample spots
Current HP [RB] - spots in current sample spots
![]() HP-X & HP-Y [RB] - spots in X and Y single samples
HP-X & HP-Y [RB] - spots in X and Y single samples
![]() HP-X & HP-Y 'sets' [RB] - spots in HP-X set and HP-Y set
HP-X & HP-Y 'sets' [RB] - spots in HP-X set and HP-Y set
![]() HP-E [RB] - spots in HPs in expression profile list
HP-E [RB] - spots in HPs in expression profile list
![]() ALL channels [RB] - ALL channels must meet the range specification
ALL channels [RB] - ALL channels must meet the range specification
![]() ANY channels [RB] - ANY channels may meet the range specification
ANY channels [RB] - ANY channels may meet the range specification
![]() AT MOST channels [RB] - AT MOST Percent SI OK channels
may meet the range specification
AT MOST channels [RB] - AT MOST Percent SI OK channels
may meet the range specification
![]() AT LEAST channels [RB] - AT LEAST Percent SI OK channels
may meet the range specification
AT LEAST channels [RB] - AT LEAST Percent SI OK channels
may meet the range specification
![]() PRODUCT of channels [RB] - the PRODUCT of all channels must meet
the range specification
PRODUCT of channels [RB] - the PRODUCT of all channels must meet
the range specification
![]() SUM of channels [RB] - the SUM of all channels must meet the
range specification
SUM of channels [RB] - the SUM of all channels must meet the
range specification
![]() Use intensity [I1:I2] sliders [CB] - use spot intensity
thresholds I1 (lower) and I2 (upper)
Use intensity [I1:I2] sliders [CB] - use spot intensity
thresholds I1 (lower) and I2 (upper)
![]() Inside [RB] - test for intensity inside of [I1:I2]
Inside [RB] - test for intensity inside of [I1:I2]
![]() Outside [RB] - test for intensity outside of [I1:I2]
Outside [RB] - test for intensity outside of [I1:I2]
![]() Use ratio [R1:R2] or Zdiff [Z1:Z2] sliders [CB] - use
spot ratio [R1:R2] or Zdiff [Z1:Z2] range thresholds
Use ratio [R1:R2] or Zdiff [Z1:Z2] sliders [CB] - use
spot ratio [R1:R2] or Zdiff [Z1:Z2] range thresholds
![]() Inside [RB] - test inside of [R1:R2] or [Z1:Z2] range
Inside [RB] - test inside of [R1:R2] or [Z1:Z2] range
![]() Outside [RB] - test outside of [R1:R2] or [Z1:Z2] range
Outside [RB] - test outside of [R1:R2] or [Z1:Z2] range
![]() Use ratio [R1:R2] or Zdiff [Z1:Z2] sliders [CB] - use
spot ratio [R1:R2] or Zdiff [Z1:Z2] range thresholds
Use ratio [R1:R2] or Zdiff [Z1:Z2] sliders [CB] - use
spot ratio [R1:R2] or Zdiff [Z1:Z2] range thresholds
![]() Inside [RB] - test inside of [R1:R2] or [Z1:Z2] range
Inside [RB] - test inside of [R1:R2] or [Z1:Z2] range
![]() Outside [RB] - test outside of [R1:R2] or [Z1:Z2] range
Outside [RB] - test outside of [R1:R2] or [Z1:Z2] range
![]() Use spot [CV] slider [CB] - apply one of the spot CV filter
modes as a Filter and popup a CV slider to set the threshold
Use spot [CV] slider [CB] - apply one of the spot CV filter
modes as a Filter and popup a CV slider to set the threshold
![]() - select
samples to be used in computing the CV
- select
samples to be used in computing the CV
![]() Use mean else max of CVs [CB] - compute the CV as the maximum
or the mean of the CVs of the samples selected
Use mean else max of CVs [CB] - compute the CV as the maximum
or the mean of the CVs of the samples selected
Filtering using statistical test by your selecting a p-value
These tests will filter genes meeting
the test criteria if the resulting p-value of that test is <= the
value specified by the p-Value state slider. Only one test may be
active at a time. If you switch to a new p-value test, it will disable
the previous p-value test. If any of these tests are selected, it
will pop up the p-Value state slider window for you to set the
p-Value. There are two t-tests: one operating on duplicate (F1,F2)
data if available, and the HP-X,HP-Y 'sets' if they are defined. The
Kolmogorov-Smirnov test operates on HP-X,HP-Y 'sets' if they are
defined. The F-test operates on the current Ordered
Condition List (OCL) consisting of any number of condition lists each
containing at least 2 (replicate) samples/condition.
![]() Filter by current Ordered
Condition List (OCL) F-Test [p-Value] slider [RB] - only
include genes that meet the F-test criteria on the current
OCL. This only works if there are at least 2 (replicate)
samples/condition for each of the condition sets in the OCL.
See info on defining
the OCL and
using the OCL data.
Filter by current Ordered
Condition List (OCL) F-Test [p-Value] slider [RB] - only
include genes that meet the F-test criteria on the current
OCL. This only works if there are at least 2 (replicate)
samples/condition for each of the condition sets in the OCL.
See info on defining
the OCL and
using the OCL data.
Filtering out genes with high replicate spot variation
The Spot CV filter mode submenu contains options to select how
the spot CV filter is to be applied. It computes the maximum value of
CV for all of the samples in the particular sample set specified. That
maximum value is then used for the spot CV filter test. Genes may be
filtered out having a large difference between spot quantification
values of corresponding duplicate spots. You may compute the
coefficient of variation CVj for the two values
(f1j and f2j for a particular
gene j.
CVj = 2|f1j-f2j|/(f1j+f2j)
If the database only has one field but replicate HPs, then you may use
the HP-X & HP-Y 'sets' CVj to filter the
genes. Then CVj values are tested against a CV
threshold slider value to eliminate genes with a high coefficient of
variation.
![]() Current HP [RB] - CV of (F1,F2) for each gene in current
sample [if duplicate spots are available on each sample]
Current HP [RB] - CV of (F1,F2) for each gene in current
sample [if duplicate spots are available on each sample]
![]() HP-X or HP-Y [RB] -
CV of (F1,F2) for HP-X and HP-Y single samples [if duplicate
spots are available on each sample]
HP-X or HP-Y [RB] -
CV of (F1,F2) for HP-X and HP-Y single samples [if duplicate
spots are available on each sample]
![]() HP-X 'set' [RB] - CV of spots in HP-X set
HP-X 'set' [RB] - CV of spots in HP-X set
![]() HP-Y 'set' [RB] - CV of spots in HP-Y set
HP-Y 'set' [RB] - CV of spots in HP-Y set
![]() HP-X or HP-Y 'sets' [RB] - CV of spots in the HP-X set or HP-Y
set
HP-X or HP-Y 'sets' [RB] - CV of spots in the HP-X set or HP-Y
set
![]() HP-X and HP-Y 'sets' [RB] - CV of spots in both the HP-X
set and HP-Y set
HP-X and HP-Y 'sets' [RB] - CV of spots in both the HP-X
set and HP-Y set
![]() HP-E [RB] - CV of HPs in expression profile list
HP-E [RB] - CV of HPs in expression profile list
2.4.3.1 Data filtering using multiple gene data filters
Any or all of the data filters may be selected simultaneously. In
particular, if you select filters that use parameter threshold
scrollers, they will be added to a state scroller window (see Figure
2.3.4.1 for details to allow adjustment of ALL sliders
simultaneously). You may change various thresholds and see the effect
in real time. Note: some of the scrollers are more sensitive to low
values. Therefore, we set them to respond non-linearly with a more
precise vernier at the low end.