4. Status and Bugs of MAExplorer
This section discusses the status and known bugs in MAExplorer. It
also discusses dealing with the reporting of fatal errors so we can
resolve them.
Section 4.1 discusses known bugs, Section 4.2
lists the revision notes for older versions known bugs. If you have experienced bugs
with an older version of MAExplorer, you might check the revision notes
to see if the bug was fixed and download a new version. Section 4.3
discusses problems in using MAExplorer
as an applet with Web browsers. Section 4.4 describes handling fatal "DRYROT" errors.
 4.1 Known Bugs in MAExplorer
4.1 Known Bugs in MAExplorer
Disclaimer: none of our code ever has bugs... :-). So despite
this, we are working on resolving these bugs and implementing planned
functionality. Here is a short non-inclusive list of known problems
that we are resolving. We welcome and encourage you to E-mail us with
any bugs that you find do exist as well as suggestions for
capabilities you would like to see. As the new open-source MAEPlugins facility evolves, most
new (and some old) functionality will migrate to these plugins. Then
the user community can help maintain these analytic methods.
If you encounter a fatal error that is detected by MAExplorer, it will
popup an error reporting
window. Please E-mail this data to us so we can try to resolve the
problem.
In the mean time, partially implemented commands are disabled to keep
you out of trouble :-) ...
You can help us and get MAExplorer to do more of the things you
would like to see. Let us know of problems that you encounter as well
as suggestions for changes or new methods you would like to see -
send us E-mail.
- 4.1.1 Browser Applet Bugs
- 4.1.2 Downloading and Installer Bugs
- 4.1.3 Computation speed and display Bugs
- 4.1.4 User state and login Status
- 4.1.5 Data file names Bug
- 4.1.6 Gene Sets Bugs
- 4.1.7 Clustering Bugs
- 4.1.9 Expression profile Bugs
- 4.1.9 Data conversion problems
- 4.1.10 Java Plugins Bugs
4.1.1 Browser Applet Bugs
- When using MAExplorer as a Web browser applet (this does not
apply if running as a stand-alone application - and we recommend
running it as a stand-alone application not as an applet), there are
problems with running MAExplorer on some of the Web browsers on some
operating systems. It generally works with Netscape 4.7 or later, on
Windows PCs (95/98/NT/2000/XP), and on SUN Unix and with Microsoft
Internet Explorer 5.0 on PCs. It may not work as well as an applet on
Macintoshes - even with Internet Explorer (although it works fine as a
stand-alone application on the Mac) and problems have been reported on
SGI systems because of browser problems.
If you are experiencing Web browser problems using the MAExplorer
applet, you might check the discussion of possible solutions.
- When run as a Web browser applet, you should only click once since multiple clicks may cause
some browsers to hang. Note that when you click the first time, it
starts downloading the applet. Clicking a second time may cause a
second request for the applet to be launched. This interferes with the
first request and so neither is started correctly.
- When the MAExplorer applet is being downloaded, some browsers
do not indicate to the user that it is being downloaded and so it
may appear that nothing is going on. Some browsers will not
indicate that it is downloading an applet. So
please be patient. When it starts, the main window will
popup and it continues to download data. Wait a while for it to load
the applet before giving up. For a 28Kb Internet connection, this
could take several minutes. When it is finished, the menu bar will
become active and it will display Ready - click on a
gene to query database.
4.1.3 Downloading and Installer Bugs
- We have had reports of problems with MAExplorer installations
or running MAExplroer on MacOS 9 since we started distributing
MAExplorer with the new InstallAnywhere version 5.0.2. We suggest you
switch to MacOS-X or use a Windows, Linux or Solaris system until this
gets resolved.
- Installer does not start or does not download the
data. Normally, when you attempt to download MAExplorer to your
computer, the InstallAnywhere software running in your Web browser
will detect which OS you are running and suggest the particular
download to use in:
"Recommended version for your computer
Download installer for ...your OS..."?
Occasionally, we have seen instances where you can not install
MAExplorer from within the Web browser. The solution is to explicitly
download the particular Platform for your OS in the Available
Installers list. And then to follow the instructions on running it.
- If you have problems downloading this with Netscape 4.7x or
later, then try Internet Explorer 5.0. It could be a Mime/type
problem with your particular browser not setup correctly.
- On Solaris, and possibly other Unix systems, you may have
problems with the stack limits. Do a "man limit" to read about the
command for your particular Unix shell. We have found that the
following seems to work. For CSH, add the following to your .cshrc
startup file. With versions 96.25 and later of MAExplorer, you may
adjust the memory size using the (Edit menu | Preferences |
Resize MAExplorer memory limits for the next time it is run) command.
This edits the startup file according to the new memory limits you
specify.
limit stacksize unlimited
4.1.4 Computation speed and display Bugs
- Because it does a lot of computation, MAExplorer runs best on
a fast computer with lots of memory. An optimal system should have at
least 128Mb of memory at least 500Mhz CPU speed. For doing
hierarchical clustering we recommend 256Mb of memory which will allow
you to cluster larger sets of genes without paging the operating
system. So you may run into problems with running out of memory on an
under-powered computer. To increase the usable memory size to more
than 256Mb (assuming your computer has it), then you may need to edit
the MAExplorer.lax file
that resides where you installed MAExplorer on your computer to
increase the heap and
stack maximum limit.
- We have seen instances where the same system will run out of
memory with particular Web browsers (again - not a problem when
running stand-alone) Netscape 4.7 on a system with a lot of
memory. This does not seem to happen with Internet Explorer 5.0. This
is because Netscape4.7 on the Windows PC seems to be able to use only
about 5.5Mb before it runs out of memory. This is not the case with
Internet Explorer 5.0.
- MAExplorer is difficult to use with a system with a small
screen because of the size and number of multiple graphic images and
plots. We recommend a screen size of at least 1024x768. As
they say these days, more is better :-), generally.
- For problems with MAExplorer and MacOS, you might also see our
MacOS problems FAQ.
- For problems with MAExplorer and Sun Solaris OS, you might
also see our SUNOS
problems FAQ.
- We have on rare occasions seen a data-related problem on MacOS
systems. If the data was edited in such a way that the line
terminators were Return characters instead of Linefeeds or
Return-Linefeeds, the data may become corrupted and MAExplorer has
difficulty reading it. The solution is to replace the Return
characters with Linefeeds or Return-Linefeeds. One (painful) way this
might be done by exporting the data to a system such as a Windows PC;
read the data into Excel; save it as an Excel worksheet file format;
exit Excel; start Excel again on the new data files; save the data
files back into the original MAExplorer directories as tab-delimited
text files. MAExplorer should recognize Return as the line
terminator. This bug will be resolved in a future release.
4.1.5 User state and login Status
- User registration and the groupware access functionality are
not available yet. However, login is available for collaborator
databases on the MGAP MAExlorer Web server.
- The commands to save and restore user states for a particular
user on the back-end server are not available yet. However, in
stand-alone mode you may save the state (including all gene and
condition sets, parameter conditions, condition sets, etc) on a local
computer by doing a (File menu | Databases | Save As DB). The
specialized groupware commands that will allow users to selectively
share their saved states with particular other users on a public
groupware server is not available yet.
4.1.6 Data file names Bug
- There is a potential problem with using long file names when
using MAExplorer with MacOS-8 or MacOS-9. Both of these operating
systems limit file names to 32 characters. This could be a problem if
importing data from another operating system (Windows, Unix, Linux,
etc) that does not have this restriction. The solution is to use short
file names. This should not be a problem with MacOS-X since it allows
file names with up to 256 characters.
4.1.7 Gene Sets Bugs
- The full range of GeneClass subsets (Section 2.4.1) is not
available yet. However, you may use the gene name guesser to find a
set of genes by wildcard into the Edited Gene List (e.g. "*ONCO*" to
find all oncogenes or proto-oncogenes, etc). These EGL sets may then
be saved as named gene sets and used in the data filter as "Filter by
'User Gene Set'". See the Gene Class ontologies
using Gene Set operations (Section 2.4.1) that can achieve a
similar effect.
4.1.8 Clustering Bugs
- The hierarchical clustering method is buggy. It currently
clusters genes - but not samples. The latter is under construction.
Centroid averaging is implemented and the arithmetic averaging is
under construction. Although it generates a clustergram plot, the
dendrogram drawing may not be drawn correctly using centroid
averaging. Depending on the number of samples, it you do a complete
hierarchical cluster for all genes and ESTs on a Netscape browser on a
Windows PC, the browser may run out of memory and nothing
appears. [When done in the stand-alone application, this is less of a
problem since there are no memory restrictions.] You can do three
things at this point: 1) switch to Internet Explorer, or 2) decrease
the number of genes being clustered (e.g. just "All named genes") or
the CV filter. The third option is to disable the clustering
cache. This will be done automatically for you and it will continue
doing the clustering. You may turn off (or on) the Use cluster
distance matrix cache in the "Hierarchical Cluster Plots
submenu. It will perform the clustering, but will take MUCH longer
without the cache.
- If you are working with very large data sets then if you start
a find similar genes or K-Means clustering with a very high threshold,
you can not change the threshold until it is done. In these
situations, try pre-setting the threshold distance, number of clusters
using the (Edit | Preferences | Adjust all Filter threshold
scrollers). This will popup the state scroller window with all of the
thresholds. You may also select the current gene prior to starting
clustering.
4.1.9 Expression profile plots
- The Expression Profile Overlay display is being improved to offer
better graphics and interaction.
- We have observed some instances of the popup EP plot where data
does not seem to correspond to the data. This may be related to the
normalization method.
- When viewing an ordered list of EP plots associated with a
sorted list of genes (e.g. clustering genes similar to a seed gene),
if there are replicated genes in the clustered genes, they may not
show up in the scrollable list of EP plots.
4.1.10 Data conversion problems
- The Cvt2Mae data file converter application is currently
available for beta-testing on the a related MAExplorer Cvt2Mae Web page. It enables users
to convert their data files (academic one-of-a-kind as well as
commercial e.g. Affymetrix, Incyte, GenePix, Scanalyze, etc) to the
standard MAExplorer files as described in Appendix C and D.
- If you are not able to convert your data using Cvt2Mae (which
is currently in early Beta testing - see the Cvt2Mae Web page for
current status), you can always manually convert the data using the
examples in Appendix C.
- We have seen some problems running the converter with some
types of data on MacOS 8-9. The temporary solution is to convert your
data using another system such as Windows until we fix the problem.
- If you are using the NCI-CIT mAdb system to package data for
MAExplorer, you must select all samples from multiple projects that
you want to compare with MAExplorer while packing the data in
mAdb. (See PDF document "How to
download mAdb data For MAExplorer") Currently, you can not easily
merge the data after you have downloaded the files to your computer.
The solution is to use mAdb to prepackage all data you want to analyze
in a single .zip file. You can not currently merge data from
separately unzipped files (without manually editing the SamplesDB.txt
file). The way you package data using mAdb is to: 1) select ALL of
the projects that contain samples that you want (on the mAdb "Top
Level Analysis Selection" web page); 2) further select the exact
subset of samples that you want in the "Array Selection" table on the
"mAdb: Database Retrieval" web page). NOTE: currently, you can only
select arrays using the same GAL file so they have compatible
geometries. This is on our list of things to change so you can mix and
match data from different chips.
4.1.11 Java Plugins bugs
- Java Plugins are in the process of being implemented and will
be made available to the user community to enable them to add their
own analysis methods to MAExplorer.
4.2 Revision notes
This section lists the revision history and is useful for deciding
whether to upgrade to the most recent release. You may want to check
for the latest the current
"Stable release" available on the MAExplorer Web site. That may be
different than the Stable
release listed in this copy of the Reference Manual. The "Beta
release" listed below the Stable release in the previous links is
experimental and may generally be downloaded as it has more
functionality. If you experience problems, you can just reinstall the
Stable version.
|
Note: An archive of some of the
stable older releases is available on the NCI/LECB Web site for a
limited period.
|
4.3 Web Browser problems when running MAExplorer as an applet
Because MAExplorer is a large system, and there may be occasional
problems running it in some Web browsers on some operating systems. We
recommend you run MAExplorer as a stand-alone application as it is
more robust.
- Normally, MAExplorer works well as an applet with Netscape 4.7
or Windows Internet Explorer 5.0 on Windows PCs and on SUN Unix. It
does not seem to work well as an applet on Macintoshs and problems
have been reported on SGI systems.
- One solution is install Sun's HotJava browser. The HotJava browser is
available from Sun Microsystems
 . You can download
HotJava for the Windows-95/-98/-NT/-2000/-XP and for Solaris
(Unix).
. You can download
HotJava for the Windows-95/-98/-NT/-2000/-XP and for Solaris
(Unix).
- Reguardless of which browser is used, because it is is doing a
lot of computation, MAExplorer runs best on a fast machine with lots
of memory. This is especially true when running a browser because of the
additional browser overhead.
4.4 Handling fatal error reporting (i.e. DRYROT errors)
If you encounter a fatal error that is detected by MAExplorer, it will
popup an error reporting window. We call this a "DRYROT" error (thanks
to "S.A.I.L." - Stanford AI Lab) because something is wrong in the
program or in the user's data files and from which it can not
recover. This type of error should not have happened. Please save and
e-mail the report to us so we can try to fix the bug or diagnose the
problem. The following figure shows an example of part of a DRYROT
error report.

Figure 4.4 Example of a fatal Dryrot Error window. This may occur for
a variety of reasons. This window lists the main reason and also lists
some of the MAExplorer state information. If you wish, you may save this
window (press the "SaveAs" button) and mail it to us. We may try to
correct the problem in the next release if it is a problem with MAExplorer.
Alternatively, it could be a user data error.
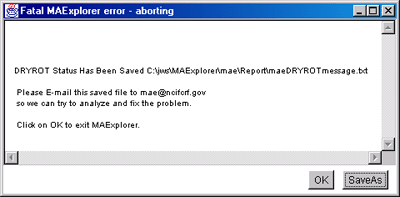
Figure 4.4.1 Example of a fatal Dryrot Error window after SaveAs.
This tells you where the saved error message file was saved and
the email address to send it to if you wish.

 4.1 Known Bugs in MAExplorer
4.1 Known Bugs in MAExplorer . You can download
HotJava for the Windows-95/-98/-NT/-2000/-XP and for Solaris
(Unix).
. You can download
HotJava for the Windows-95/-98/-NT/-2000/-XP and for Solaris
(Unix).


